Can you study expression levels using northern blot? The answer is a resounding “yes!” This article explores the basics of northern blot, including the invariant control mRNA, Probes, Detection, and Comparison with other methods. For more information, read on! Then, use the information to answer your research questions. Alternatively, you can find more information on how to do this method from other sources.
Invariant control mRNA
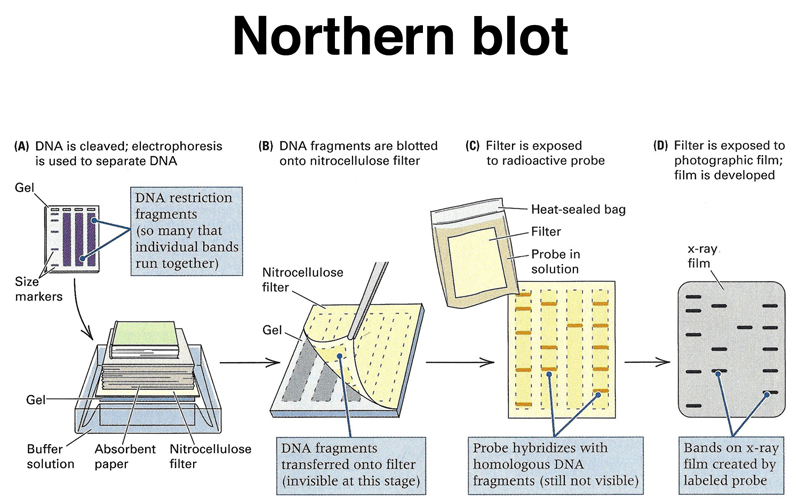
Northern blot analysis is the logical first step in characterization of mRNA expression. It allows the visualization of mRNAs in their intact forms, while also providing information on their size. In addition, this method allows for great flexibility, as it does not require specific cloning vectors or primers. However, in some applications, the flexibility of the Northern blot is compromised by its reliance on nonspecific hybridization.
Northern blot methods rely on the use of widely expressed mRNAs. Commonly used standards include b-actin, a-tubulin, b2-microglobulin, glyceraldehyde-3-phosphate dehydrogenase, and hypoxanthine-guanine phosphoribosyl transferase. Additionally, probes specific for 18S and 28S ribosomal RNA (rRNA) are also used.
Probes
Probes for studying expression levels using northern blet have three different functions. In one case, a probe is an RNA molecule that binds to the blot to measure the level of the gene. The probe may be radioactive or contain a bound enzyme. In the other case, it can simply be a molecule that is designed to hybridize with a cellular RNA sequence.
Using a RNA or DNA probe in Northern blot analysis is the preferred method for determining the amount of a particular gene’s expression in a sample. Alternatively, oligonucleotide probes can also be used. Probes are synthesised using random-primed labeling or radiolabeled oligonucleotide strands. The size of the band will be dependent on the probe’s specific activity. Generally, high specific activities are used to detect low levels of RNA, while low-specific activity probes are suitable for detection of abundant RNAs.
Detection
A Northern blot detects the expression of specific gene transcripts. The probes used for Northern blot detection typically contain either a full or partial cDNA sequence. They may be labeled with radioactive nucleotides, chemiluminescent probes, or haptens. The resulting signal is then detected using autoradiography. The signals are then compared to a standard curve to determine the relative abundance of the target mRNA.
The sensitivity of Northern blot analysis is limited. Low-level transcription may not be detected. One example of such transcription is the case with GI-GPx mRNA. However, Western blotting showed that the mRNA was expressed in rat liver, and RT-PCR confirmed this finding. For more information, you can visit the Northern blot page at Wikimedia Commons.
Comparison with other methods
Northern blots have the advantage of being able to compare the relative abundance of mRNA species but are not useful for absolute quantitation. Northern blots require that each sample on the blot be hybridized with either the mRNA of interest or a standard probe which is present in equal amounts in all samples. This hybridization can be carried out concurrently with the experimental probes or after the blot is stripped of its original mRNA.
Reverse northern analysis was a novel method. This technique was previously uncommon but has since evolved into gene expression profiling, which monitors the levels of a wide range of genes. This method can be applied to many kinds of experiments, including plant and animal cell culture, RNAi analysis, and gene expression profiling. Northern blots are commonly used to measure gene expression levels and detect the expression of many genes in different tissues.
Limitations
The Northern blotting method is a widely used technique for analyzing gene expression levels. However, it has several limitations. One is that it uses relatively large amounts of RNA, which can be expensive for large-scale analyses. Another is that the method is susceptible to RNase degradation, which may require careful optimization and use of sterile materials. This article describes the main advantages of Northern blotting and discusses the limitations of the method.
Northern blots are often published on online databases. BlotBase contains nearly 700 published Northern blots from 25 different tissue types, and a searchable database lets you search by blot ID and gene identifier. Once you find a blot that matches your research question, you can view its image and read more information about the gene expression level. By using an online database, you can share your findings with other researchers, which is a useful feature.
Applications
In 1977, the Northern blot was introduced for RNA analysis. Named after DNA Southern blot, Northern blotting uses denaturing gel electrophoresis and blotting to detect specific target RNAs. Although this technique lacks the sensitivity and specificity of fluorescence-based qPCR, it remains useful for studies of gene expression or RNA degradation. In addition to its use for detection of mRNA, Northern blots can also be used for analyzing RNA degradation. However, the Northern blot can lead to nonspecific hybridization which can confound data interpretation. Therefore, it is important to use radioactivity when interpreting the data. Another useful method is the RNase protection assay, which can be used to identify and characterize different RNA species.
Northern blots have several advantages. It allows researchers to monitor the expression levels of a wide variety of genes. However, the technique is laborious, especially when several probes are required. First, one must remove the first probe and then add a second one. This will compromise the accuracy of subsequent rounds. Secondly, Northern blots only measure steady-state mRNA accumulation, not RNA stability or transcription rates.